
About
Comprehensive atlas for regulatory elements and functional annotations
Dataset Available
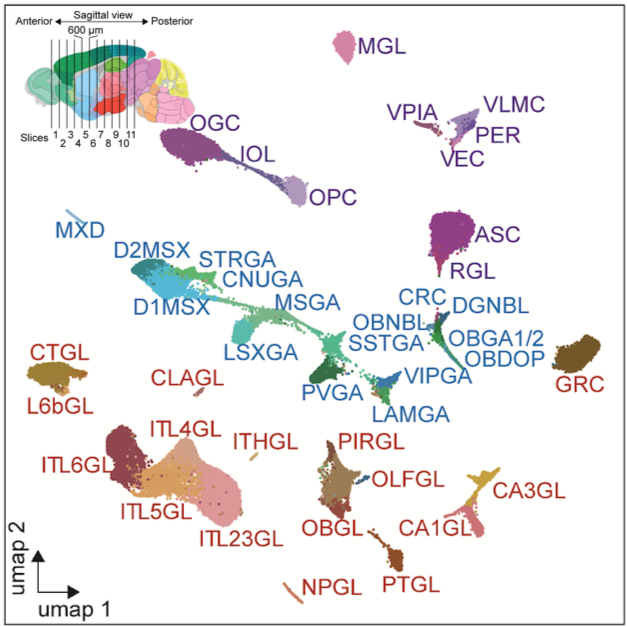
Mouse Brain
Species: Mus musculus Sample: 45 brain sub-regions Method: snATAC-seq Nuclei count: > 800,000
Last updated May,2020
Explore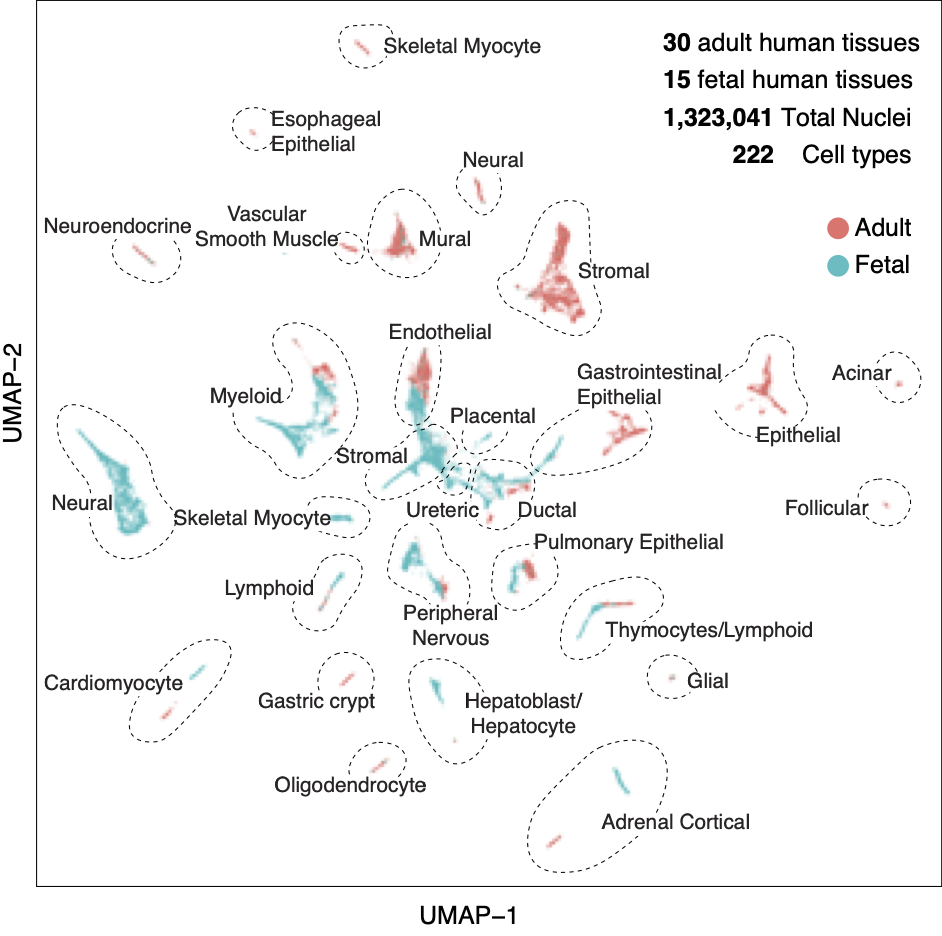
Human Tissues
Species: Homo SapiensSamples: 30 adult tissues Method: snATAC-SeqCell counts: >1.3 million
Explore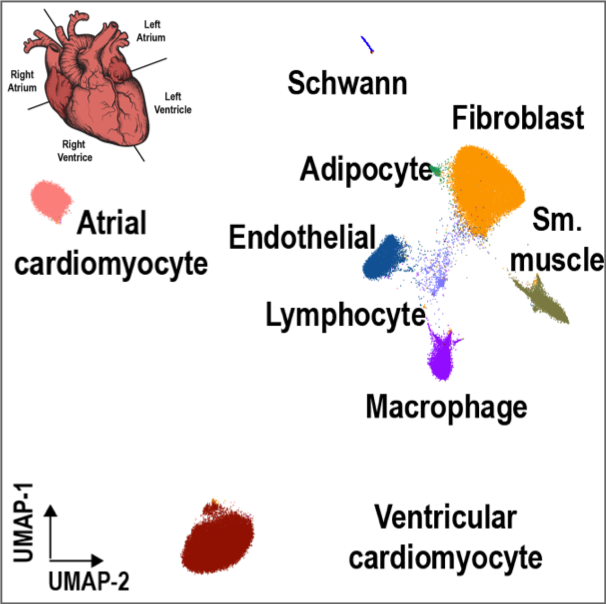
Human Heart
Species: Homo Sapiens Samples: Left/right Atria and ventricules from 4 donors Method: snATAC-Seq and snRNA-Seq Cell counts: 80K snATAC cells and 36K snRNA cells
Explore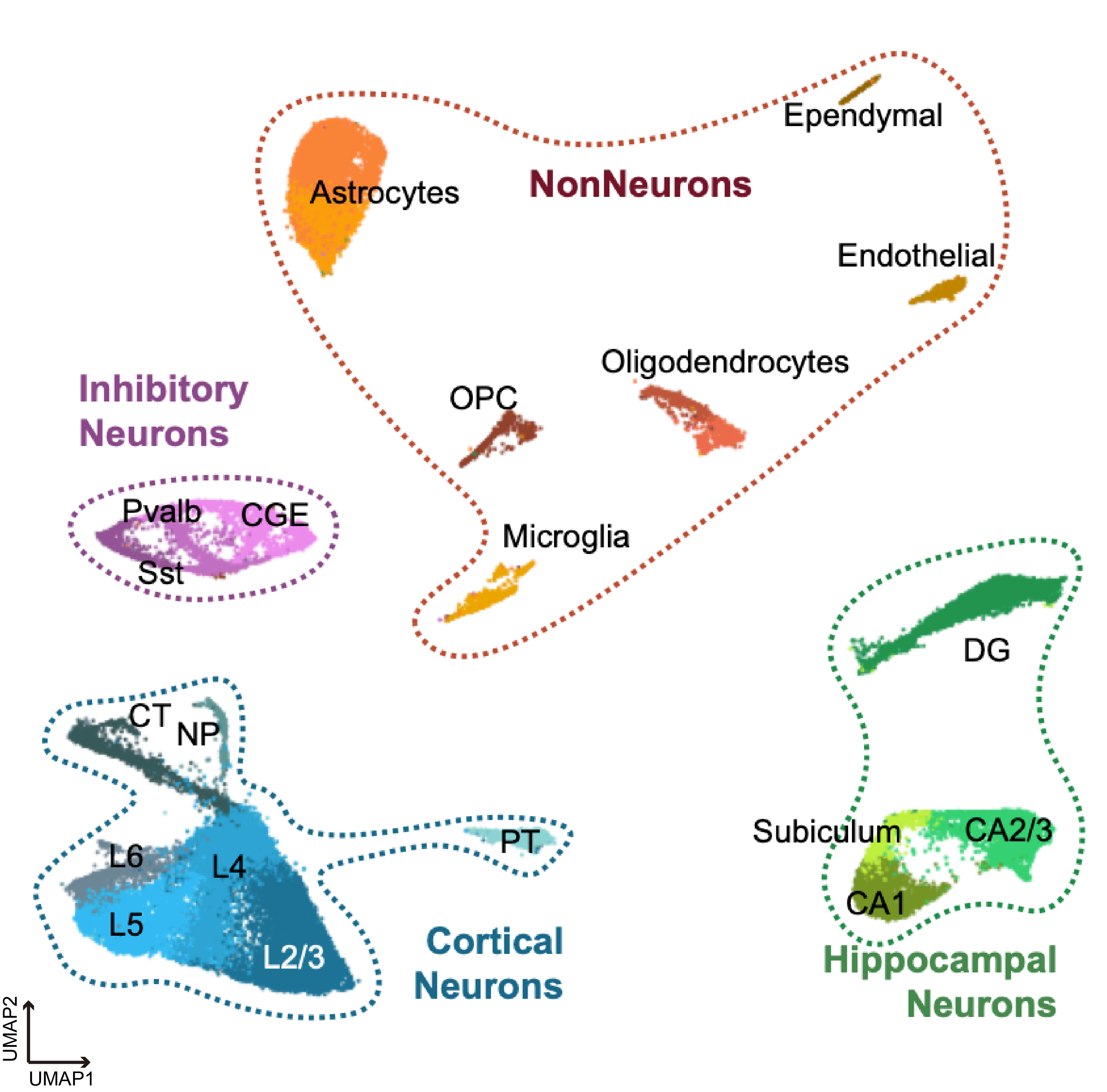
Mouse Cortex
Species: Mus musculusSamples: Frontal cortex & hippocampus Method: Paired-TagCell counts: 64,849
Explore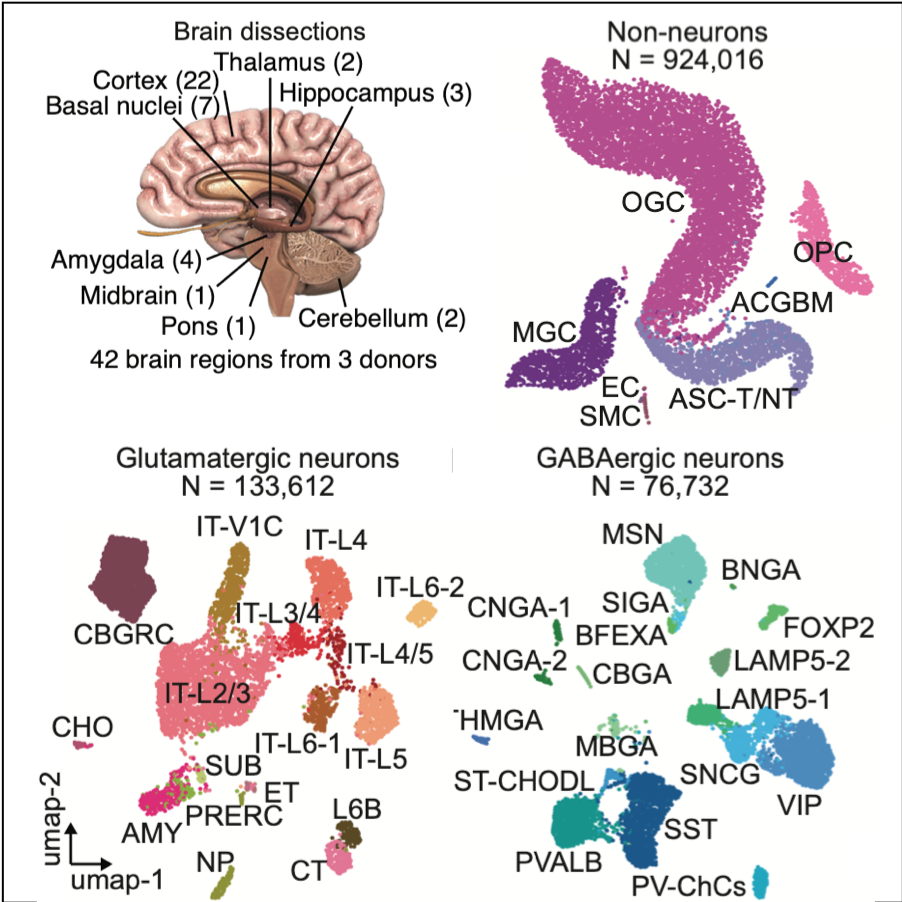
Human Brain
Species: Homo SapiensSamples: 42 brain regions Method: snATAC-seqCell counts: >1.1 million
Explore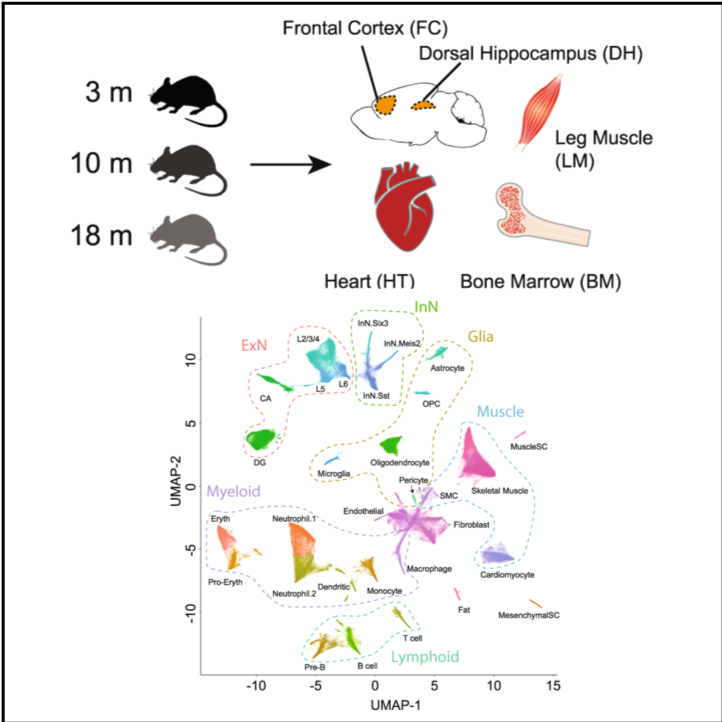
Mouse Aging
Species: Mus musculusSamples: brains, hearts, skeletal muscles, and bone marrows from young, middle-aged, and old miceMethod: snATAC-seqCell counts: 227,529
Explore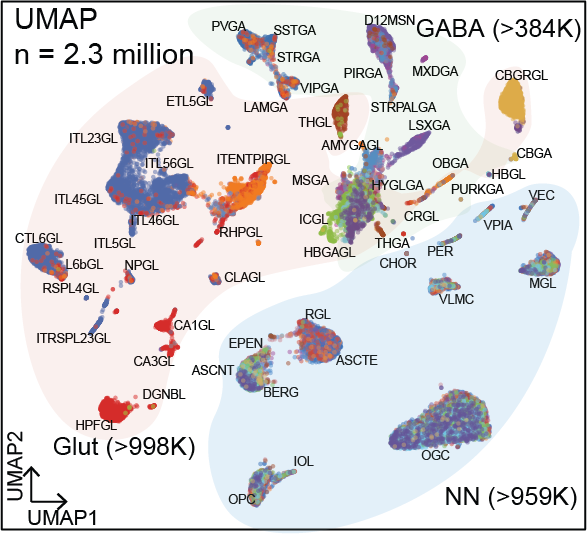
Whole Mouse Brain
Species: Mus musculus Sample: 117 brain sub-regions Method: snATAC-seq Nuclei count: > 2.3 million
Last updated March,2023
Explore
Coming soon
About
We hope you find this data resource useful. Please write us with experiences and suggestions. We would love stories about how people are using it in their work.
Summary
We provide comprehensive reference maps of cis-regulatory elements identified from single-cell epigenetic datasets. You can explore cell clusters from different tissues and organisms, detailed annotations for every cell types, and genomic signal tracks and functional predictions.
Publications
2022
A comparative atlas of single-cell chromatin accessibility in the human brain
Yang Eric Li, Sebastian Preissl, Michael Miller, Nicholas D Johnson, Zihan Wang, Henry Jiao, Chenxu Zhu, Zhaoning Wang, Yang Xie, Olivier Poirin, Colin Kern, Antonio Pinto-Duarte, Wei Tian, Kimberly Siletti, Nora Emerson, Julia Osteen, Jacinta Lucero, Lin Lin, Qian Yang, Sarah Espinoza, Quan Zhu, Nathan Zemke, Anna Marie Yanny, Julie Nyhus, Nick Dee, Tamara Casper, Nadiya Shapovalova, Daniel Hirschstein, Rebecca D Hodge, Sten Linnarsson, Trygve E Bakken, Boaz Levi, C Dirk Keene, Jingbo Shang, Ed S Lein, Allen Wang, M Margarita Behrens, Joseph R Ecker, Bing Ren. bioRxiv, 2022. doi: https://doi.org/10.1101/2022.11.09.515833
Single-cell Epigenome Analysis Reveals Age-associated Decay of Heterochromatin Domains in Excitatory Neurons in the Mouse Brain
Yanxiao Zhang*,#, Maria Luisa Amaral*, Chenxu Zhu, Steven F. Grieco, Xiaomeng Hou, Lin Lin, Justin Buchanan, Liqi Tong, Sebastian Preissl, Xiangmin Xu#, Bing Ren#. Cell Research, 2022. doi:10.1038/s41422-022-00719-6
2021
A single-cell atlas of chromatin accessibility in the human genome
Kai Zhang*, James D. Hocker*, Michael Miller, Xiaomeng Hou, Joshua Chiou, Olivier B. Poirion, Yunjiang Qiu, Yang E. Li, Kyle J. Gaulton, Allen Wang, Sebastian Preissl, Bing Ren. Cell, 2021. doi:https://doi.org/10.1016/j.cell.2021.10.024
An Atlas of Gene Regulatory Elements in Adult Mouse Cerebrum
Yang Eric Li*, Sebastian Preissl*, Xiaomeng Hou, Ziyang Zhang, Kai Zhang, Yunjiang Qiu, Olivier Poirion, Bin Li, Joshua Chiou, Hanqing Liu, Antonio Pinto-Duarte, Naoki Kubo, Xiaoyu Yang, Rongxin Fang, Xinxin Wang, Jee Yun Han, Jacinta Lucero, Yiming Yan, Michael Miller, Samantha Kuan, David Gorkin, Kyle Gaulton, Yin Shen, Michael Nunn, Eran A. Mukame, M. Margarita Behrens, Joseph Ecker and Bing Ren. Nature, 2021. doi:https://doi.org/10.1038/s41586-021-03604-1
Cardiac Cell Type-Specific Gene Regulatory Programs and Disease Risk Association
James D Hocker, Olivier B Poirion, Fugui Zhu, Justin Buchanan, Kai Zhang, Joshua Chiou, Tsui-Min Wang, Qingquan Zhang, Xiaomeng Hou, Yang Eric Li, Yanxiao Zhang, Elie Farah, Allen Wang, Andrew D McCulloch, Kyle J Gaulton, Bing Ren#, Neil C Chi#, Sebastian Preissl# Science Advances, 2021. doi:10.1126/sciadv.abf1444
Joint profiling of histone modifications and transcriptome in single cells from mouse brain
Chenxu Zhu, Yanxiao Zhang, Yang Eric Li, Jacinta Lucero, M. Margarita Behrens & Bing Ren. Nature Methods, 2021. doi:10.1038/s41592-021-01060-3
2018
Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation
Sebastian Preissl*, Rongxin Fang*, Hui Huang, Yuan Zhao, Ramya Raviram, David U. Gorkin, Yanxiao Zhang, Brandon C. Sos, Veena Afzal, Diane E. Dickel, Samantha Kuan, Axel Visel, Len A. Pennacchio, Kun Zhang, Bing Ren. Nature Neuroscience, 2018. doi:10.1038/s41593-018-0079-3
Contributes




Contact
-
Lab location: 9500 Gilman Dr, CMME #2071, La Jolla, CA-92093, USA (Ludwig Institute for Cancer Research, San Diego) Lab phone: +1-858-822-5767 Fax: +1-858-534-7750 Web Maintenance: Yang (Eric) Li < yal054 AT health.ucsd.edu > Bin Li < bil022 AT ucsd.edu >
